Splicing, Exons, and the SMN2 ‘Back-Up’ Gene
Splicing, Exons, and the SMN2 ‘Back-Up’ Gene
Read our text report below. You can also watch this YouTube video produced in 2016 by Cold Spring Harbour Laboratory with Youreka Science:
Proteins such as the SMN protein are made by cells from genes found in our DNA (Figure 1). First, DNA must be copied into a second similar molecule called messenger ribonucleic acid (mRNA). mRNA is then used as a template to produce proteins.
The individual components of a protein (amino acids) and the order in which they are put together is dictated by the individual components of the mRNA (nucleotides), which are dependent on the building blocks of the DNA (also nucleotides). In this way, information in our genes affects how our proteins are put together.
Before mRNA, which at this point is called pre-mRNA, is converted to protein, there is a special process called “splicing” that must occur.
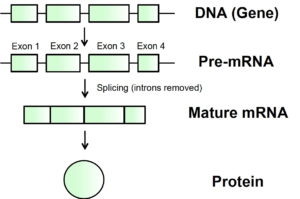
Figure 1. From DNA to protein. Stretches of DNA known as genes are copied into pre-mRNA with a corresponding sequence. Before pre-mRNA can be used as a template for protein production, the exons, which encode important information about the protein, must be spliced or stuck together. Between exons, regions known as introns are found that do not contain information necessary to build a protein. Introns are removed from pre-mRNA in the process of splicing to produce mature mRNA, which can then be translated into protein.
The pre-mRNA includes important sections called exons, which contain the information to produce parts of a protein. Remember, exon 7 is often missing from the SMN protein produced by the SMN2 gene.
These exons are interspersed by regions called introns that do not contain important information for the protein sequence. These introns are removed from pre-mRNA, and the remaining exons are spliced, or stuck, back together to produce mature mRNA ready to be used as a template for protein production.
SMN2 and Splicing
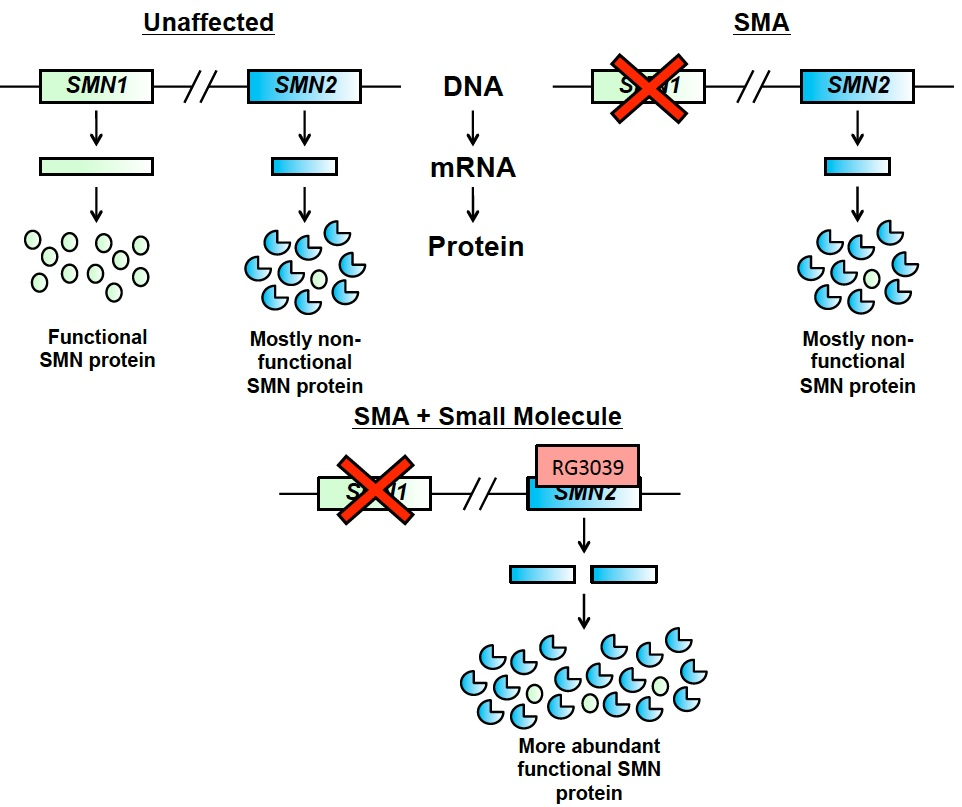
Humans possess two genes that code for and make SMN protein: SMN1 and SMN2. SMN1 produces fully functional, full-length SMN protein, and is the gene that when mutated leads to SMA. SMN2, however, due to a single difference in the DNA, only produces approximately 10% of the amount of SMN protein that is made from SMN1.
This single difference between the two SMN genes results in exon 7 of SMN2 to be mis-spliced out of the mature RNA and thus not incorporated into 90% of the SMN protein made by the ‘back-up’ gene. This protein lacking exon 7 is not able to perform the normal function of SMN. This is not a problem in people with a working copy of the SMN1 gene. However, in people lacking functioning SMN1, this poses an issue because insufficient SMN protein is produced, leading to SMA.
Nevertheless, SMN2 has emerged as a potential therapeutic target for SMA. This is because if expression from SMN2 can be increased or the single difference in the DNA between it and SMN1 can be corrected, more SMN protein will be produced, which has the potential to reduce disease severity.
Antisense oligonucleotides (ASOs), such as nusinersen, work by targeting the messenger RNA of SMN2 and causing more efficient inclusion of exon 7 (Figure 2). This leads to a greater percentage of the protein made by SMN2 being functional.
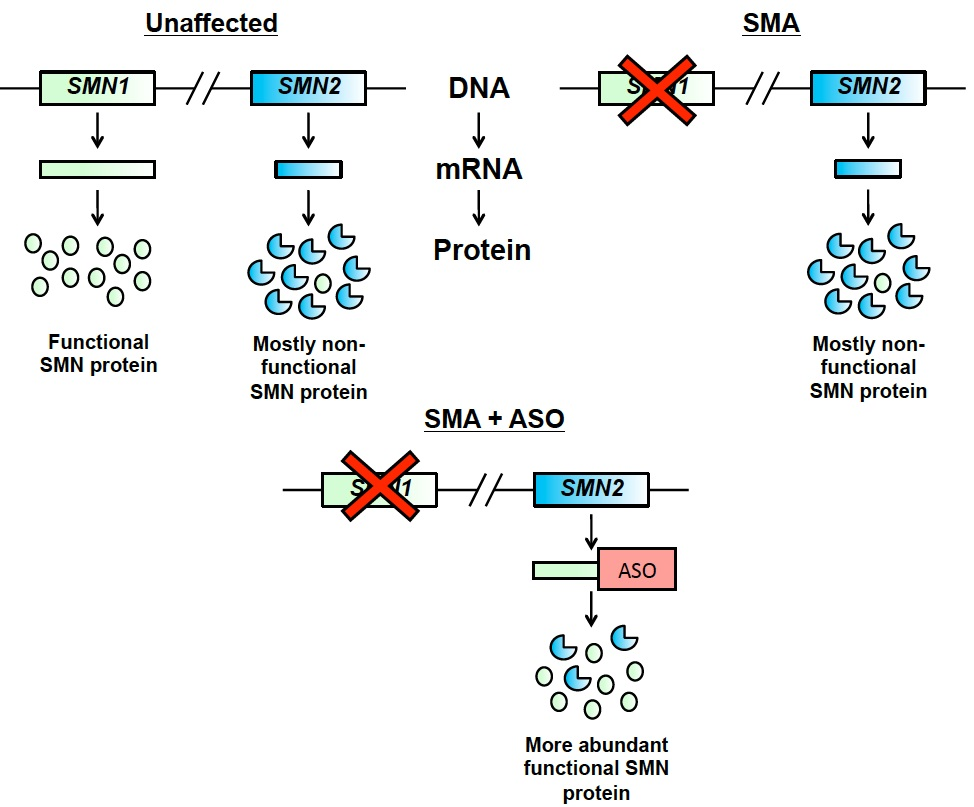
Figure 2. Antisense oligonucleotides target SMN2 mRNA to increase exon 7 inclusion, and therefore increase the amount of functional SMN protein produced by the SMN2 gene.
Rather than acting on the mRNA, certain small molecule chemicals, such as RG3039, are able to target the DNA of SMN2 and increase the amount of total protein it produces (Figure 3). In doing so, these compounds are able to increase the amount of functional SMN protein produced by the SMN2 gene.