Splicing and the SMN2 ‘Back-Up’ Gene
Splicing and the SMN2 ‘Back-Up’ Gene
Proteins are made by cells of the body using our DNA. Genes are short and specific stretches of DNA that contain the information needed to make a particular protein (Figure 1). For example, the SMN1 gene provides the necessary details to make the SMN protein.
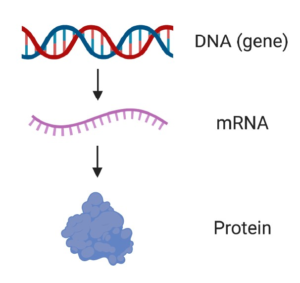
Figure 1. From DNA to mRNA to protein. A gene is a specific piece of DNA that contains the information needed to make a particular protein (e.g. the SMN1 gene is used to make the SMN protein). mRNA is an intermediate molecule that is copied from DNA and used as a template to make protein.
DNA is made up of four different types of molecule called nucleotides (Figure 2). The order in which these nucleotides are found provides the specific set of instructions to make a particular protein.
The individual building blocks of proteins are called amino acids (Figure 2). The instructions about which amino acids to include in a protein are found in the sequence of nucleotides within our genes.
Changes in the the order or number of the nucleotides within a gene can affect the protein that is made, and are known as mutations. Mutations in our DNA can cause genetic diseases.
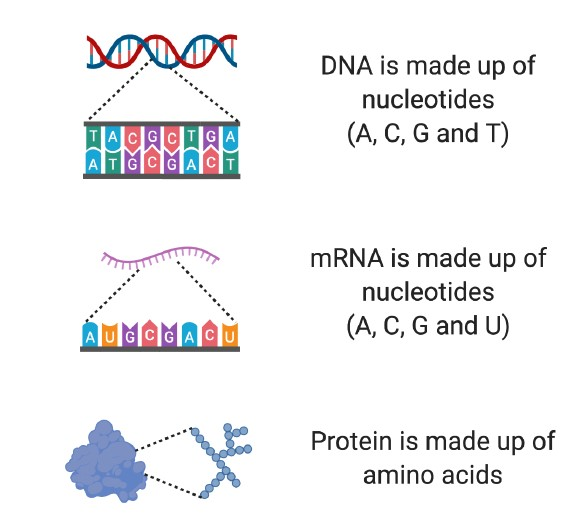
Figure 2. Building blocks of DNA, mRNA and protein. DNA is made up of four different nucleotides (A, C, G and T). mRNA is made up of four different nucleotides, three of which are the same as found in DNA (A, C, G and U). Proteins are made from amino acids.
Before a protein can be made using our DNA, the DNA must first be copied into a similar molecule called mRNA, which serves as a template from which the protein is made (Figure 1). Because the mRNA is copied from the DNA, it provides the instructions to make a protein that is specific for the gene that was copied.
To make things a bit more complicated, which is important to understanding how some drugs with potential for SMA actually work, the mRNA can be divided into two main categories: pre-mRNA and mature mRNA (Figure 3).
When mRNA is first made from DNA, it is known as pre-mRNA. pre-mRNA is a direct copy of the DNA and has two major components: exons and introns. Exons are the most important sections of the pre-mRNA, because they contain the information needed to produce parts of a protein. Several exons are usually found in pre-mRNA (See the example in Figure 3, which has three exons).
Between exons are parts of the pre-mRNA called introns, which do not contain important information for making the protein.
Before a protein can be made, the introns must be removed in a process called splicing. Introns are removed from pre-mRNA, and the remaining exons are spliced, or stuck, back together to produce mature mRNA, which is then used to make protein.
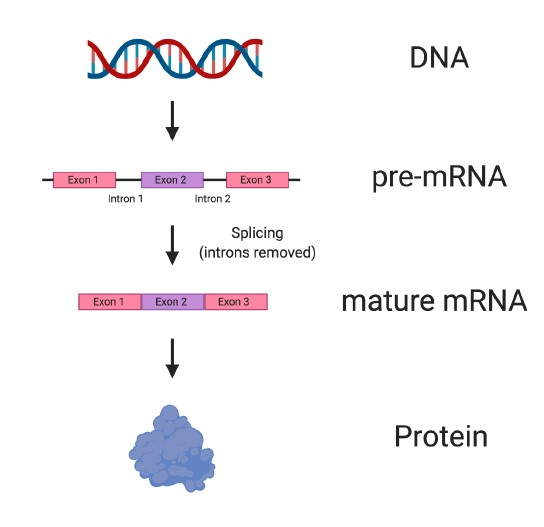
Figure 3. Splicing converts pre-mRNA to mature RNA. Stretches of DNA known as genes are copied into pre-mRNA with a corresponding sequence. Before pre-mRNA can be used to make protein, the exons, which contain important information about the protein, must be spliced or stuck together. Between exons, regions known as introns are found, which do not contain information needed to make a protein. Introns are removed from pre-mRNA in a process called splicing, which results in mature mRNA being made that can then be used to make protein.
In humans, two genes can make the SMN protein: SMN1 and SMN2. SMN1 produces full-length, functional SMN protein (Figure 4), and is the gene that when missing or mutated leads to SMA.
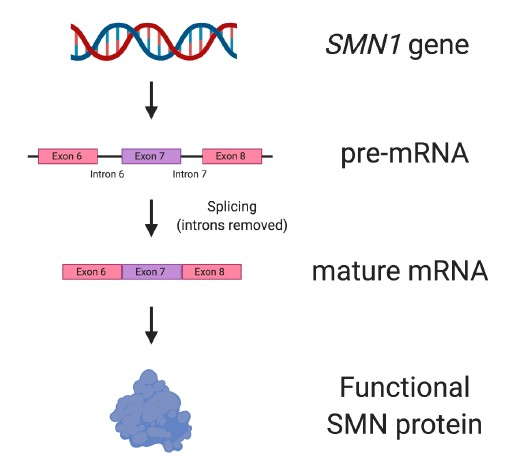
Figure 4. The SMN1 gene. The SMN1 gene produces all the SMN protein needed for motor neurons to survive and be healthy. SMN1 has nine exons, but not all them are depicted in the figure.
Due to a single nucleotide difference between the SMN1 and SMN2 genes, the SMN2 ‘back-up’ gene produces only approximately 10% of the amount of SMN protein that is made from SMN1 (Figure 5).
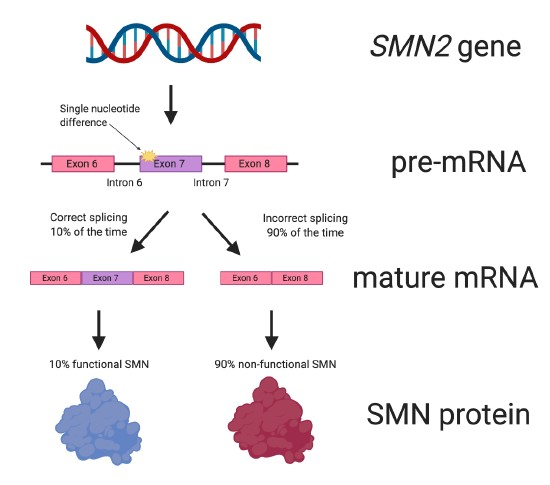
Figure 5. The SMN2 gene. Due to a single nucleotide difference from the SMN1 gene, the SMN2 ‘backup’ gene only produces about 10% of the amount of functional SMN protein that SMN1 can make. SMN2 has nine exons, but not all them are depicted in the figure.
This difference between the two SMN genes affects the splicing of SMN2. When SMN2 pre-mRNA is spliced and the mature mRNA is formed (see Figure 3 for a reminder), about 90% of the mature RNA lacks the important exon 7 (Figure 5). The SMN protein made from the mature mRNA lacking exon 7 does not work properly because it is missing important amino acids.
This is not a issue in people with a working copy of the SMN1 gene, because SMN1 produces enough SMN protein by itself. However, in people lacking a working SMN1 gene, this causes a problem because not enough SMN protein is made, leading to SMA.
SMN2 has been used as a potential target for SMA treatment for many years. This is because, unlike SMN1, SMN2 is present in all people with SMA. If the production of SMN protein from SMN2 can be increased in SMA, it has the potential to reduce disease severity.
This strategy is how Spinraza / nusinersen is able to increase the amount of SMN protein, leading to its approval as a treatment for SMA.
Spinraza is an antisense oligonucleotide, which means that it is a small stretch of synthetic DNA (i.e. made in the laboratory) capable of recognising the nucleotide sequence of part of a specific mRNA.
Spinraza works by targeting the pre-mRNA of the SMN2 gene (Figure 6). It specifically binds to a portion of intron 7. This causes exon 7 to be included more frequently in the mature mRNA.
Put another way, Spinraza improves the efficiency of SMN2 splicing. This ultimately results in more SMN protein being made from the SMN2 gene.
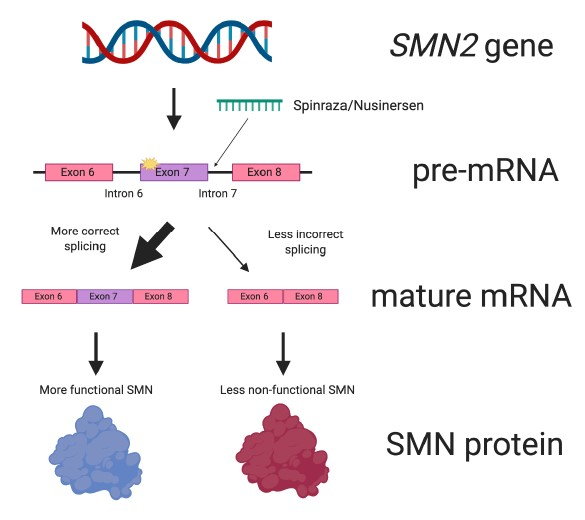
Figure 6. Spinraza works by improving splicing of SMN2. Spinraza is a small molecular patch that specifically interacts with SMN2 pre-mRNA. Doing so increases the amount of mature mRNA that contains the all-important exon 7. This leads to more SMN protein being made.
Roche’s small molecule drug, Risdiplam, also interacts with SMN2 pre-mRNA to enhance exon 7 inclusion and thus increase SMN protein levels. Because Risdiplam affects SMN2 splicing, it is often called a “splice-modifying” or “splicing modulator” drug. The Novartis experimental therapy, Branaplam, (since discontinued in 2021) also worked in a similar manner.
At the International Scientific Congress on SMA in March 2024, the following update was provided:
Ilaria Signoria (University Medical Centre, Utrecht, Netherlands), a PhD student working with Ludo Van Der Pol and Ewout Groen, presented her work aiming to find out why there is a wide range of responses in the clinic to the SMN-dependent treatments of nusinersen and risdiplam.
On a population level, the more copies of SMN2 a person has, the less severe the SMA is likely to be. However, on an individual level, this is not always true; for example, people with SMA Type 2 may have two, three or four copies of SMN2. Similarly, when SMN-dependent treatments (e.g. nusinersen and risdiplam) are given to people with the same SMA Type and number of SMN2 gene copies, the effect on symptoms can differ between recipients.
Unfortunately, we do not currently know the reason for these difference between SMA subtypes and between treatment responses.
To provide some insight into this, Ms. Signoria grew skin cells that had been collected from 35 people with SMA (Types 1 to 4) and ten unaffected controls. Each SMA skin sample was taken from people with known SMN genetics and a detailed history of their condition. The skin cells, known as ‘fibroblasts’, are easily obtained, grow and multiply very quickly, and have the same genetic makeup as that of the donor; hence, they are a useful tool for assessing the impact of SMN-dependent therapies in the laboratory.
The skin cells from all 45 people were separately grown and assessed. The shape and size of all cells was evaluated, as were the levels of SMN protein. In addition, the intermediate molecule that is created from DNA to make a protein, known as ‘messenger RNA’, was also assessed – see sections above for more info.
There were no clear differences in the shape and size of the skin cells from people with SMA compared to controls. However, as expected, there were reductions in the SMN protein and the ‘messenger RNA’ from the SMN1 and SMN2 genes in the SMA ‘fibroblasts’.
Skin cells taken from people with more SMN2 copies generally had higher availability of SMN protein and ‘messenger RNA’. Furthermore, those cells with more SMN2 copies tended to produce more SMN protein when treated with the SMN2-targeting therapies nusinersen and risdiplam.
Interestingly, it was found that the levels of SMN ‘messenger RNA’ did not correlate well with the amount of SMN protein, suggesting that factors involved in converting the ‘messenger RNA’ into protein differ between individuals, and that the process can be more efficient in some people than others.
It was also observed that factors such as age, SMN2 copy number and SMN levels before treatment were only able to explain a proportion of the variability in response to the treatments. This suggests that other currently unknown factors are at play.
The ‘fibroblast’ model is a powerful tool to identify what these factors are. This is exciting because, in the future, we may be able to manipulate these factors in people with SMA to improve individual responses to the SMN-dependent therapies.
